George Tsiolas
Research Fellow | Molecular Biology | Applied Bioinformatics
Institute of Applied Biosciences, CERTH
About me
As a Molecular Biologist, I have hands-on experience in NGS, bioinformatics, and genome editing. I am proficient in using various bioinformatics tools and programming languages like R and Bash scripting. I have a solid background in genomic and transcriptomic research and can tackle any analytical challenge that comes my way. Additionally, I am a great communicator and enjoy collaborating in teams. My skills include NGS, bioinformatics, genome editing, programming languages such as R and Bash scripting, genomic and transcriptomic research, analytical problem-solving, communication, and teamwork.
In addition, I have recent experience in the start-up ecosystem. I founded μBio, a start-up that tackled the problem of AMR in Greek aquacultures. Although μBio couldn’t survive, this experience has given me a unique perspective on the challenges and opportunities of working in a start-up environment. Furthermore, my experience in the start-up ecosystem has changed my perspective on applied research. I have come to appreciate the importance of translating scientific discoveries into practical solutions that can benefit society. I am now more motivated than ever to apply my skills and knowledge to real-world problems and make a positive impact on people’s lives.
- Entrepreneurship
- Open Innovation
- Applied Bioinformatics
- Molecular Biomimetics
M.Sc. in Biomedical and Molecular Sciences in Diagnosis and Treatment, 2016 - 2018
Medical Department, Democritus University of Thrace
B.Sc. in Molecular Biology and Genetics, 2009 - 2014
Department of Molecular Biology and Genetics, Democritus University of Thrace
Skills
100 %
95 %
95 %
85 %
100 %
100 %
Experience
Responsibilities include:
- Experimental Design
- Performing Experiments and Analyses
- Publications and Deliverables
Classes include:
- Real-Time PCR methods to detect adulteration
- 16S Nanopore sequencing to reveal Feta cheece microbiome
Projects
Featured Publications
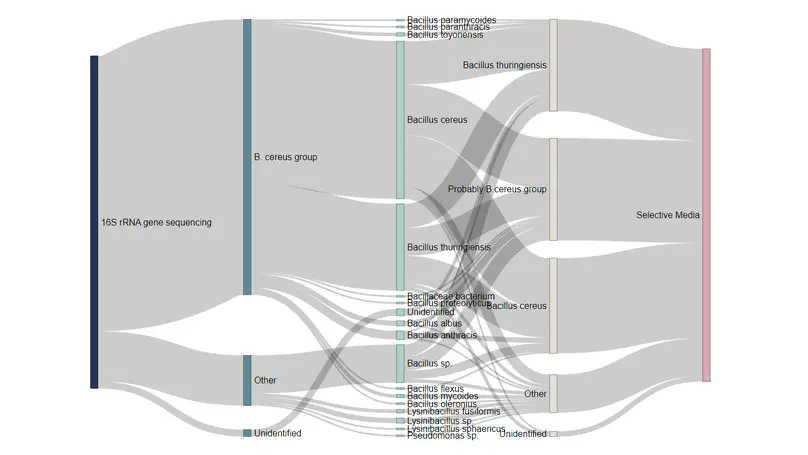
Bacillus cereus is a spore-forming, facultative anaerobic bacterium, well known for its ability to cause food poisoning and spoilage of milk and dairy products. Identification of B. cereus in contaminated milk can be laborious and time-consuming. The current widely used method for the isolation and identification of B. cereus includes cultivation in mannitol egg yolk polymyxin (MYP) agar, forming pink-purple colonies surrounded by a pink halo of egg-yolk precipitate (lecithinase positive). PCR methods targeting the 16S rRNA gene have also been established for identifying B. cereus in milk products, and can be more sensitive and rapid.
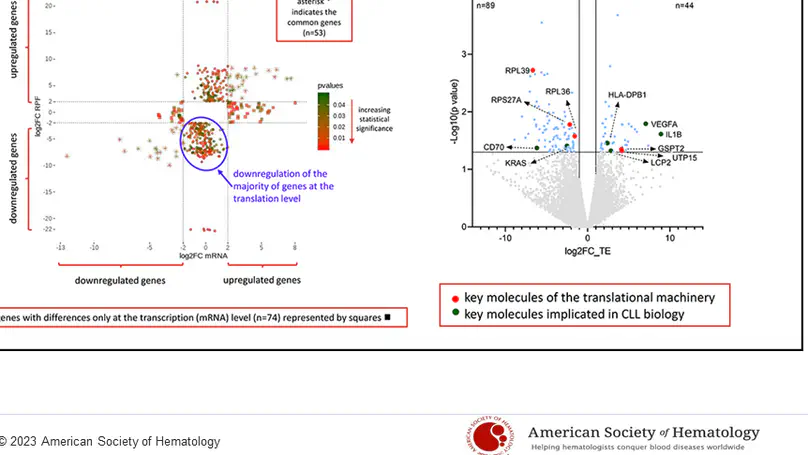
Recent studies of chronic lymphocytic leukemia (CLL) have reported recurrent mutations in the RPS15 gene, which encodes the ribosomal protein S15 (RPS15), a component of the 40S ribosomal subunit. Despite some evidence about the role of mutant RPS15 (mostly obtained from the analysis of cell lines), the precise impact of RPS15 mutations on the translational program in primary CLL cells remains largely unexplored. Here, using RNA sequencing and ribosome profiling, a technique that involves measuring translational efficiency, we sought to obtain global insight into changes in translation induced by RPS15 mutations in CLL cells. To this end, we evaluated primary CLL cells from patients with wildtype or mutant RPS15 as well as MEC1 CLL cells transfected with mutant or wild-type RPS15. Our data indicate that RPS15 mutations rewire the translation program of primary CLL cells by reducing their translational efficiency, an effect not seen in MEC1 cells. In detail, RPS15 mutant primary CLL cells displayed altered translation efficiency of other ribosomal proteins and regulatory elements that affect key cell processes, such as the translational machinery and immune signaling, as well as genes known to be implicated in CLL, hence highlighting a relevant role for RPS15 in the natural history of CLL.
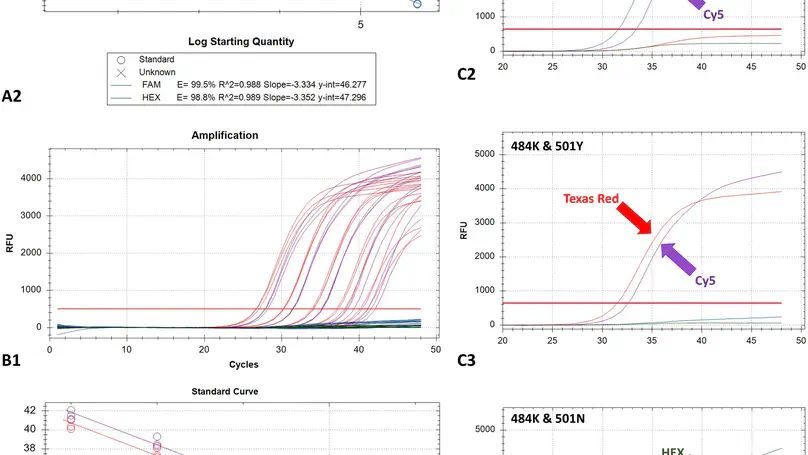
The emergence of SARS-CoV-2 mutations resulting in the S protein amino-acid substitutions N501Y and E484K, which have been associated with enhanced transmissibility and immune escape, respectively, necessitates immediate actions, for which their rapid identification is crucial. For the simultaneous typing of both of these mutations of concern (MOCs), a one-step real-time RT-PCR assay employing four locked nucleic acid (LNA) modified TaqMan probes was developed. The assay is highly sensitive with a LOD of 117 copies/reaction, amplification efficiencies >94 % and a linear range of over 5 log10 copies/reaction. Validation of the assay using known SARS-CoV-2-positive and negative samples from human and animals revealed its ability to correctly identify wild type strains, and strains possessing either one or both targeted amino-acid substitutions, thus comprising a useful pre-screening tool for rapid MOC identification. The basic principles of the methodology for the development of the assay are explained in order to facilitate the rapid design of similar assays able to detect emerging MOCs.
All Publications
Contact
I’d love to hear from you, whether you have a question, feedback, or just want to say hello! Drop me a message using the form below, and I’ll get back to you as soon as possible. Thank you for visiting my website!
- george.tsiolas@certh.gr
- +30 2311 257 521
- 6th km Harilaou-Thermis Road, Thessaloniki, Central Macedonia 57001
- INAB building at CERTH campus
- Monday to Friday, 9:00 to 17:00
- Book an appointment